Post-processing¶
Functions:
|
Adds the additional coordinates along each dataset dimension as new columns in the factor matrices. |
|
Convert a factor matrix into a tidy dataset, for use with Plotly Express. |
|
Label the CP tensor by converting the factor matrices into DataFrames with a sensible index. |
|
Standard postprocessing of a CP decomposition. |
|
Resolve the sign indeterminacy of CP models. |
- tlviz.postprocessing.add_factor_metadata(cp_tensor, dataset)[source]¶
Adds the additional coordinates along each dataset dimension as new columns in the factor matrices.
The coordinates of xarray DataArrays can contain metadata. For each dimension, there may be additional coordinates that are not used for indexing purposes. This function will iterate over all modes of a dataset and a labelled CP tensor and add the additional coordinates as new columns in the factor matrices.
- Parameters:
- cp_tensorlabelled CP Tensor
- datasetxarray.DataArray
- Returns:
- tuple
CP-tensor like tuple where the factor matrices are augmented with additional metadata.
Examples
>>> from tlviz.data import load_oslo_city_bike >>> from tlviz.postprocessing import postprocess, add_factor_metadata >>> from tensorly.decomposition import parafac >>> bikes = load_oslo_city_bike() >>> bikes.coords Coordinates: * End station name (End station name) object '7 Juni Plassen' ... 'Økernve... lat (End station name) float64 59.92 59.93 ... 59.93 59.92 lon (End station name) float64 10.73 10.75 ... 10.8 10.78 * Hour (Hour) int32 0 1 2 3 4 5 6 7 8 ... 16 17 18 19 20 21 22 23 * Month (Month) int32 1 2 3 4 5 6 7 8 9 10 11 12 * Day of week (Day of week) int32 0 1 2 3 4 5 6 * Year (Year) int32 2020 2021
We see that the
End station namedimension has two additional columns:latandlon. These contain metadata about the end station coordinates, and it can be useful to have these columns also in the factor matrices. To do this, we first fit a PARAFAC model to the dataset, then we postprocess it to label the CP tensor and finally, we add the metadata information>>> cp = parafac(bikes.data, 3, init="random") >>> cp_labelled = postprocess(cp, bikes) >>> print(cp_labelled[1][0].columns) RangeIndex(start=0, stop=3, step=1) >>> cp_with_metadata = add_factor_metadata(cp_labelled, bikes) >>> print(cp_with_metadata[1][0].columns) Index([0, 1, 2, 'lat', 'lon'], dtype='object')
We see that when we add the metadata, then the latitude and longitude columns are added to the dataframe.
- tlviz.postprocessing.factor_matrix_to_tidy(factor_matrix, var_name='Component', value_name='Signal', id_vars=None)[source]¶
Convert a factor matrix into a tidy dataset, for use with Plotly Express.
If we convert a factor matrix into a tidy dataset (or long table), then we get a table on the form
Tidy format factor matrix¶ Index
Component
Signal
0
0
0.1
1
0
0.5
…
…
…
38
2
0.7
39
2
0.2
The component vectors are all stacked on top of each other, with a separate column that specifies which component each row belongs to. This function can also preserve metadata, which is signified by columns that have non-integer column names. For example, if we have a dataframe on the form
Factor matrix with metadata¶ Index
0
1
2
lat
lon
0
0.1
0.2
0.5
59.91273
10.74609
1
0.5
0.2
0.1
63.43049
10.39506
…
…
…
…
…
…
5
0.2
0.1
0.3
60.39299
5.32415
5
0.0
0.2
0.1
58.97005
5.73332
and convert it into a tidy format factor matrix, then we get a table on the form
Tidy format factor matrix with metadata¶ Index
lat
lon
Component
Signal
0
59.91273
10.74609
0
0.1
1
63.43049
10.39506
0
0.5
…
…
…
…
…
4
69.6489
18.95508
2
0.0
5
58.97005
5.73332
2
0.1
- Parameters:
- factor_matrixpd.DataFrame
A labelled factor matrix potentially with metadata columns
- var_namestr
Name of the column that specifies which component each row belongs to
- value_namestr
Name of the column that holds the magnitude of each component entry
- id_varsiterable or None (default=None)
Which columns to interpret as metadata. The columns not specified here are considered as the components. If
id_vars is None, then all columns with non-integer names are considered metadata columns.
- Returns:
- pd.DataFrame
Tidy format factor matrix
Examples
>>> import numpy as np >>> import pandas as pd >>> from tlviz.postprocessing import factor_matrix_to_tidy >>> rng = np.random.default_rng(0) >>> factor_matrix = pd.DataFrame(rng.uniform(size=(10, 3))) >>> factor_matrix.head() 0 1 2 0 0.636962 0.269787 0.040974 1 0.016528 0.813270 0.912756 2 0.606636 0.729497 0.543625 3 0.935072 0.815854 0.002739 4 0.857404 0.033586 0.729655 >>> tidy_factor_matrix = factor_matrix_to_tidy(factor_matrix) >>> tidy_factor_matrix.head() index Component Signal 0 0 0 0.636962 1 1 0 0.016528 2 2 0 0.606636 3 3 0 0.935072 4 4 0 0.857404 >>> factor_matrix_with_metadata = factor_matrix.copy() >>> factor_matrix_with_metadata["Metadata"] = rng.uniform(size=10) >>> factor_matrix_with_metadata.head() 0 1 2 Metadata 0 0.636962 0.269787 0.040974 0.688447 1 0.016528 0.813270 0.912756 0.388921 2 0.606636 0.729497 0.543625 0.135097 3 0.935072 0.815854 0.002739 0.721488 4 0.857404 0.033586 0.729655 0.525354 >>> tidy_factor_matrix_with_metadata = factor_matrix_to_tidy(factor_matrix_with_metadata) >>> tidy_factor_matrix_with_metadata.head() Metadata index Component Signal 0 0.688447 0 0 0.636962 1 0.388921 1 0 0.016528 2 0.135097 2 0 0.606636 3 0.721488 3 0 0.935072 4 0.525354 4 0 0.857404
- tlviz.postprocessing.label_cp_tensor(cp_tensor, dataset)[source]¶
Label the CP tensor by converting the factor matrices into DataFrames with a sensible index.
Convert the factor matrices into Pandas DataFrames where the DataFrame indices are given by the coordinate names of an xarray DataArray. If the dataset has only two modes, then it can also be a pandas DataFrame.
- Parameters:
- cp_tensorCPTensor
CP Tensor whose factor matrices should be labelled
- datasetxarray.DataArray of pandas.DataFrame
Dataset used to label the factor matrices
- Returns:
- CPTensor
Tuple on the CPTensor format, except that the factor matrices are DataFrames.
- tlviz.postprocessing.postprocess(cp_tensor, dataset=None, reference_cp_tensor=None, permute=True, resolve_mode=None, unresolved_mode=-1, flip_method='transpose', weight_behaviour='normalise', weight_mode=0, allow_smaller_rank=False, include_metadata=False)[source]¶
Standard postprocessing of a CP decomposition.
This function will perform standard postprocessing of a CP decomposition. If a reference CP tensor is provided, then the columns of
cp_tensor’s factor matrices are aligned with the columns ofreference_cp_tensor’s factor matrices.Next, the factor matrices of the CP tensor are scaled according the the specified weight behaviour (default is normalised).
If a dataset is provided, then the sign indeterminacy is resolved and if the dataset is labelled (i.e. is an xarray or a dataframe), then the factor matrices of the CP tensor is labelled too.
This function is equivalent to calling
permute_cp_tensordistribute_weightsresolve_cp_sign_indeterminacylabel_cp_tensor
- Parameters:
- cp_tensorCPTensor or tuple
CPTensor to postprocess
- datasetndarray or xarray (optional)
Dataset the CP tensor represents
- reference_cp_tensorCPTensor or tuple (optional)
If provided, then the tensor whose factors we align the CP tensor’s columns with.
- permutebool
If
True, then the factors are permuted in descending weight order ifreference_cp_tensorisNone.- resolve_modeint, iterable or None
Mode(s) whose factor matrix should be aligned with the data. If None, then the sign should be corrected for all modes except the
unresolved_mode.- unresolved_modeint
Mode used to correct the sign indeterminacy in other mode(s). The factor matrix in this mode may not be aligned with the data.
- method“transpose” or “positive_coord”
Which method to use when computing the signs. Use
"transpose"for the method in [BAK08, BLJ13], and"positive_coord"for the method corrected for non-orthogonal factor matrices described above.- weight_behaviour{“ignore”, “normalise”, “evenly”, “one_mode”} (default=”normalise”)
How to handle the component weights.
"ignore"- Do nothing"normalise"- Normalise all factor matrices"evenly"- All factor matrices have equal norm"one_mode"- The weight is allocated in one mode, all other factor matrices have unit norm columns.
- weight_modeint (optional)
Which mode to have the component weights in (only used if
weight_behaviour="one_mode")- allow_smaller_rankbool (default=False)
If
True, then a low rank decomposition can be permuted against one with higher rank. The “missing columns” are padded by nan values- include_metadatabool (default=True)
If
True, then the factor metadata will be added as columns in the factor matrices.
- Returns:
- CPTensor
The post processed CPTensor.
See also
Examples
Here is an example were we use postprocess on a decomposition of aminoacid data
>>> import tlviz >>> import numpy as np >>> import matplotlib.pyplot as plt >>> from tensorly.decomposition import parafac >>> dataset = tlviz.data.load_aminoacids() Loading Aminoacids dataset from: Bro, R, PARAFAC: Tutorial and applications, Chemometrics and Intelligent Laboratory Systems, 1997, 38, 149-171
The dataset is an xarray DataArray and it contains relevant side information
>>> print(type(dataset)) <class 'xarray.core.dataarray.DataArray'>
We see that after postprocessing, the cp_tensor contains pandas DataFrames
>>> cp_tensor = parafac(dataset.data, 3, init="random", random_state=0) >>> cp_tensor_postprocessed = tlviz.postprocessing.postprocess(cp_tensor, dataset) >>> print(type(cp_tensor[1][0])) <class 'numpy.ndarray'> >>> print(type(cp_tensor_postprocessed[1][0])) <class 'pandas.core.frame.DataFrame'>
We see that after postprocessing, the factor matrix has unit norm
>>> print(np.linalg.norm(cp_tensor[1][0], axis=0)) [160.82985402 182.37338941 125.3689186 ] >>> print(np.linalg.norm(cp_tensor_postprocessed[1][0], axis=0)) [1. 1. 1.]
When we construct a dense tensor from a postprocessed cp_tensor it is constructed as an xarray DataArray
>>> print(type(tlviz.utils.cp_to_tensor(cp_tensor))) <class 'numpy.ndarray'> >>> print(type(tlviz.utils.cp_to_tensor(cp_tensor_postprocessed))) <class 'xarray.core.dataarray.DataArray'>
The visualisation of the postprocessed cp_tensor shows that the scaling and sign indeterminacy is taken care of and x-xaxis has correct labels and ticks
>>> fig, ax = tlviz.visualisation.components_plot(cp_tensor) >>> plt.show()
(Source code, png, hires.png, pdf)
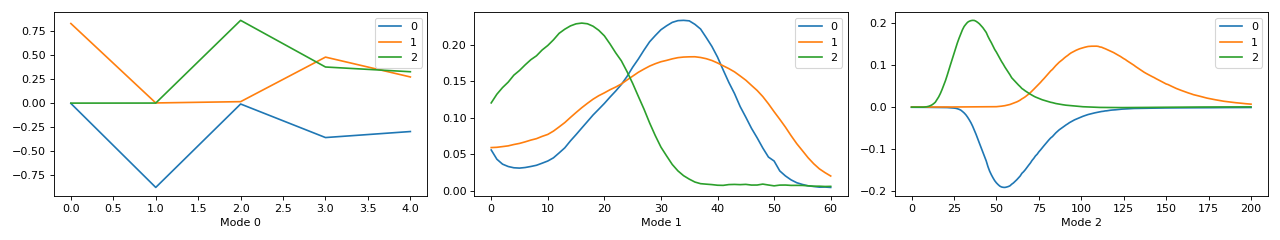
>>> fig, ax = tlviz.visualisation.components_plot(cp_tensor_postprocessed) >>> plt.show()
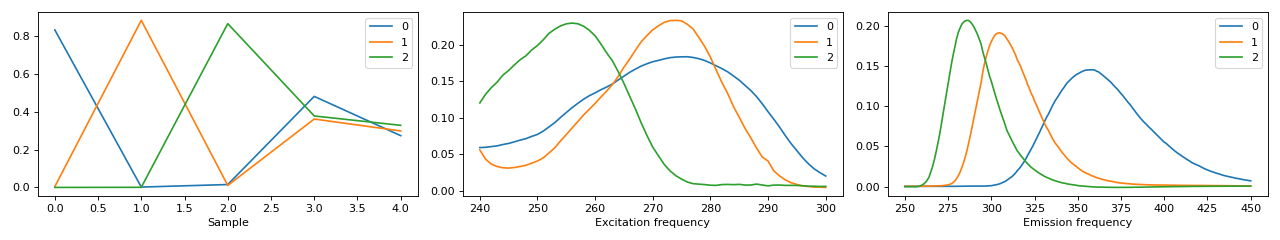
- tlviz.postprocessing.resolve_cp_sign_indeterminacy(cp_tensor, dataset, resolve_mode=None, unresolved_mode=-1, method='transpose')[source]¶
Resolve the sign indeterminacy of CP models.
Tensor factorisations have a sign indeterminacy that allows any change in the sign of the component vectors in one mode, under the condition that the sign of a component vector in another mode changes as well. This means that we can “flip” any component vector so long as the corresponding component vector in another mode is also flipped. This flipping can hurt the model’s interpretability. For example, if a factor represents a chemical spectrum, then this flipping may lead to it being negative instead of positive.
To illustrate the sign indeterminacy, we start with the SVD, which is on the form
\[\mathbf{X} = \mathbf{U} \mathbf{S} \mathbf{V}^\mathsf{T}.\]The factorisation above is equivalent with the following factorisation:
\[\mathbf{X} = (\mathbf{U} \text{diag}(\mathbf{f})) \mathbf{S} (\mathbf{V} \text{diag}(\mathbf{f}))^\mathsf{T},\]where \(\mathbf{f}\) is a vector containing only ones or negative ones. Similarly, a CP factorisation with factor matrices \(\mathbf{A}, \mathbf{B}\) and \(\mathbf{C}\) is equivalent to the CP factorisations with the following factor matrices:
\((\mathbf{A} \text{diag}(\mathbf{f})), (\mathbf{B} \text{diag}(\mathbf{f}))\) and \(\mathbf{C}\)
\((\mathbf{A} \text{diag}(\mathbf{f})), \mathbf{B}\) and \((\mathbf{C} \text{diag}(\mathbf{f}))\)
\(\mathbf{A}, (\mathbf{B} \text{diag}(\mathbf{f}))\) and \((\mathbf{C} \text{diag}(\mathbf{f}))\)
One way to circumvent the sign indeterminacy is by imposing non-negativity. However, that is not always a reasonable choice (e.g. if the data also contains negative entries). When we don’t want to impose non-negativity constraints, then we need some other way to resolve the sign indeterminacy (which this function provides). The idea is easiest described in the two-way (matrix) case.
Consider a data matrix, \(\mathbf{X}\) whose columns represent samples and rows represent measurements. Then, we want the measurement-mode component-vectors to be mostly aligned with the data matrix. The components should describe what the data is, not what it is not. For example, if the data is non-negative, then the measurement-mode component vectors should be mostly non-negative. With the SVD, we can compute whether we should flip the \(r\)-th column of \(\mathbf{U}\) by computing
\[f_r = \sum_{i=1^I} v_{ir}^2 \text{sign}{v_{ir}}\]if \(f_r\) is negative, then we should flip the sign of the \(r-th\) column of \(\mathbf{U}\) and \(\mathbf{V}\) [BAK08].
The methodology above works well in practice, and is rooted in the fact that the \(i\)-th row of \(\mathbf{V}\) can be interpreted as the coordinates of the \(i\)-th row of \(\mathbf{X}\) in a vector space spanned by the columns of \(\mathbf{U}\). Then, the above equation will give us component vectors where the data points is mainly located in the non-negative orthant.
The above interpretation is correct under the assumption: \(\mathbf{U}^\mathsf{T}\mathbf{U} = \mathbf{I}\). However, the heuristic still works well when this is not the case [BLJ13]. Still, we also include a modification of the above scheme where the same interpretation holds with non-orthogonal factors.
\[f_r = \sum_{i=1}^I h_{ir}^2 \text{sign}(h_{ir}),\]where \(\mathbf{H} = \mathbf{U}(\mathbf{U}^\mathsf{T}\mathbf{U})^{-1} \mathbf{X}\). That is the rows of \(\mathbf{H}\) represent the rows of \(\mathbf{X}\) as described by the column basis of \(\mathbf{U}\).
In the multiway case, when \(\mathcal{X}\) is a tensor instead of a matrix, we can apply the same logic [BLJ13]. If we have the factor matrices \(\mathbf{A}, \mathbf{B}\) and \(\mathbf{C}\), then we flip the sign of any factor matrix (e.g. \(\mathbf{A}\)) by computing
\[f_r^{(\mathbf{A})} = \sum_{i=1}^I {h_{ir}^{(\mathbf{A})}}^2 \text{sign}({h_{ir}^{(\mathbf{A})}}),\]where \(\mathbf{H}^{(\mathbf{A})} = \mathbf{A}^\mathsf{T} \mathbf{X}_{(0)}\) or \(\mathbf{H}^{(\mathbf{A})} = \mathbf{A}(\mathbf{A}^\mathsf{T}\mathbf{A})^{-1} \mathbf{X}_{(0)}\), depending on whether the scheme based on the SVD scheme [BLJ13] or the corrected scheme. \(\mathbf{X}_{(0)} \in \mathbb{R}^{I \times JK}\) is the tensor, \(\mathcal{X}\), unfolded along the first mode. We can then correct the sign of \(\mathbf{A}\) by multiplying and one of the other factor matrices by \(\text{diag}(\mathbf{f}^{(\mathbf{A})})\). By using this procedure, we can align all factor matrices except for one (the unresolved mode) with the “direction of the data”.
Note that this sign indeterminacy comes as a direct consequence of the scaling indeterminacy of component models, since \(\text{diag}(\mathbf{f})^{-1} = \text{diag}(\mathbf{f})\).
- Parameters:
- cp_tensorCPTensor or tuple
TensorLy-style CPTensor object or tuple with weights as first argument and a tuple of components as second argument.
- resolve_modeint, iterable or None
Mode(s) whose factor matrix should be aligned with the data. If None, then the sign should be corrected for all modes except the
unresolved_mode.- unresolved_modeint
Mode used to correct the sign indeterminacy in other mode(s). The factor matrix in this mode may not be aligned with the data.
- method“transpose” or “positive_coord”
Which method to use when computing the signs. Use
"transpose"for the method in [BAK08, BLJ13], and"positive_coord"for the method corrected for non-orthogonal factor matrices described above.
- Returns:
- CPTensor or tuple
The CP tensor after correcting the signs.
- Raises:
- ValueError
If
unresolved_modeis not between-dataset.ndimanddataset.ndim-1.- ValueError
If
unresolved_modeis inresolve_mode
Notes